Research
Our research broadly aims to uncover mechanisms that enable cells to identify and resolve disruptions during protein synthesis.
Mechanisms that enable cells restore proteostasis following translational stress
Current Projects
Mechanisms regulating the Ribotoxic Stress Response during translational stress
Mechanisms regulating translational repression through the Integrated Stress Response pathway during ribotoxic stress
Overview
Cells must respond to diverse external and internal perturbations to ensure survival. These responses are coordinated by various signaling and quality control (QC) pathways that converge on gene expression and stress response programs to regulate cell fate. Proliferating cells, which allocate up to 30% of their energy to proteostasis, rely on ribosomal health and function for their overall fitness. As such, pathogenic infections, environmental insults, proteostasis defects, and metabolic disorders that damage ribosomes or disrupt protein synthesis activate various stress response and adaptive pathways.
Elongation distress is a key trigger for regulation
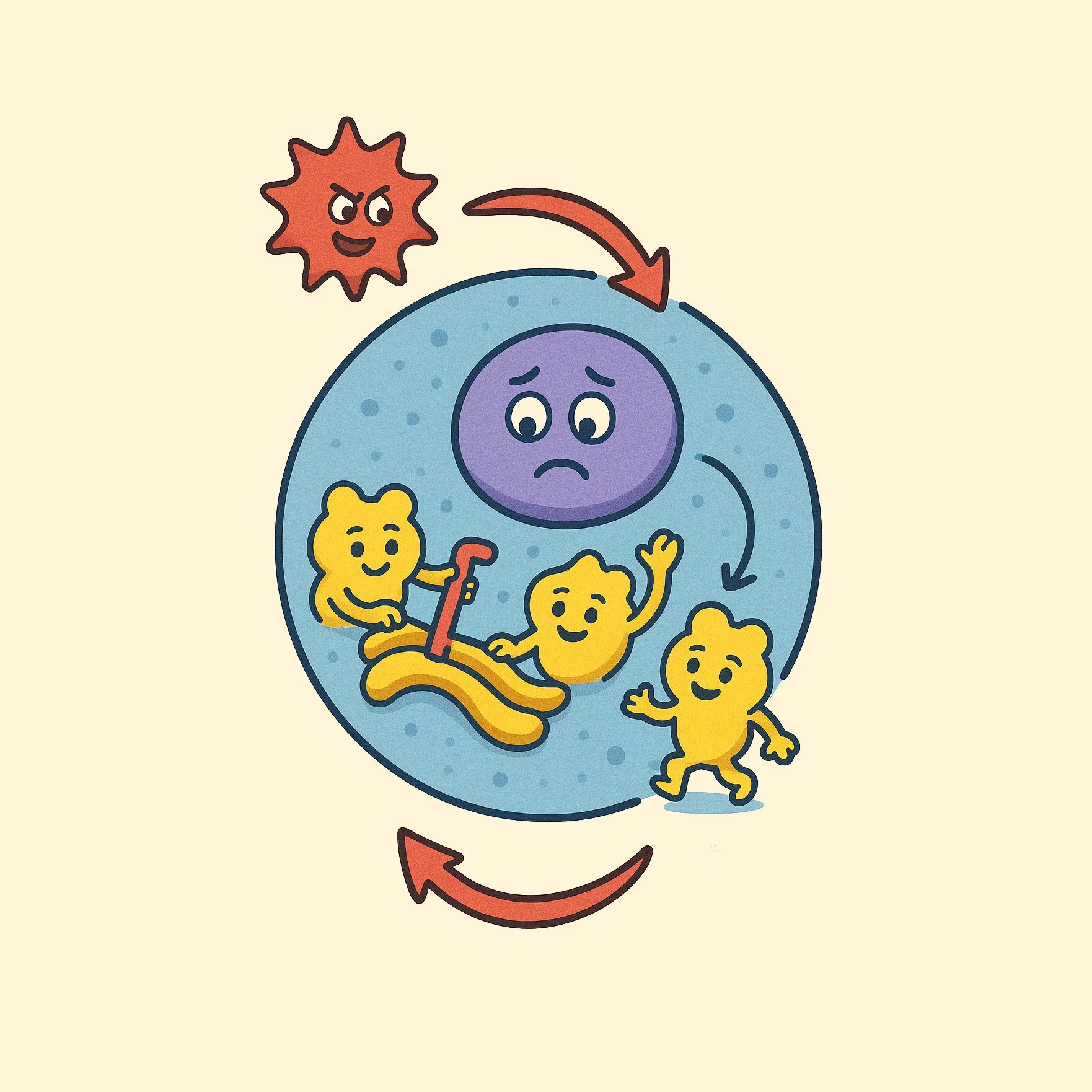
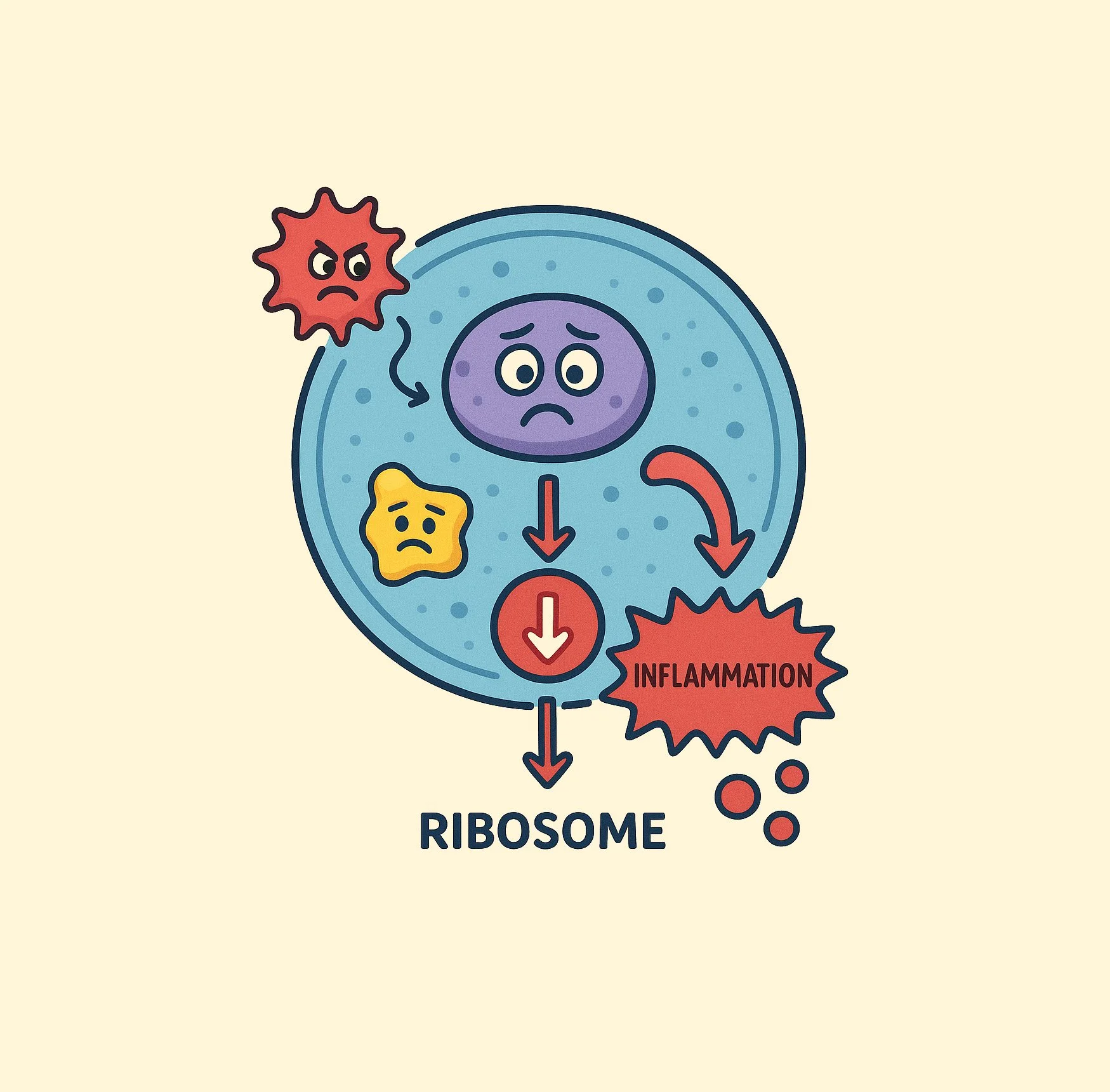
We use a combination of biochemical, proteomic, genetic, and structural approaches to identify and characterize regulators of ribosome-mediated stress responses. In previous work, we discovered a QC factor called EDF1 that acts as an “early responder” during ribosomal collisions and plays a central role in restoring translational homeostasis (Sinha et al., eLife 2020). More recently, we discovered that the ribotoxic stress response, rather than the DNA damage response, plays a dominant role in determining cell fate outcomes following UV exposure (Sinha et al., Cell 2024).
Our research broadly aims to uncover mechanisms that enable cells to identify and resolve disruptions during protein synthesis. We focus on ribosomal collisions, which occur when stalled ribosomes are rear-ended on damaged mRNAs during faulty elongation. While collisions have been recognized as crucial for detecting aberrant translation, the key regulators of various ribosome-mediated QC and signaling pathways remain poorly characterized.
EDF1 is an early responder during ribosomal collisions (adapted from Sinha et al., 2020)
ZAK regulates cell fate responses according to the level and duration of ribosome collisions. (adapted from Sinha et al., 2024)
This pathway, regulated by the protein kinase ZAK, is activated in response to ribosomal collisions and determines whether a cell will survive or undergo apoptosis based on the extent of ribosomal and RNA damage. Our findings established ribosomes as physiological sensors of UV-induced cellular damage, playing pivotal roles in determining cellular fate (Sinha et al., Cell 2024). Our long-term goal is to harness these mechanisms to target inherent vulnerabilities in cells that display unusually high protein synthesis rates, such as cancer cells, as well as in aging cells with abnormalities in proteostasis and protein turnover.
Mechanisms regulating the Ribotoxic Stress Response during translational stress
ZAK is the principal MAP3 kinase surveilling ribosomal health in cells. We seek to understand how ribosomal crowding modulates ZAK's activity by driving its transition from an autoinhibited to activated state. Our work will focus on characterizing how various co-factors and the phosphorylation status of ZAK collectively regulate its activation dynamics, oligomerization, localization, substrate selectivity, deactivation, and stability. By defining these regulatory mechanisms, we aim to uncover how ZAK activation thresholds adapt to varying translational demands across cell types. These insights will help identify ZAK-centric vulnerabilities in cancers and other proliferative contexts.
Mechanisms regulating ZAK activity in cells
Mechanisms regulating translational control in cancer cells
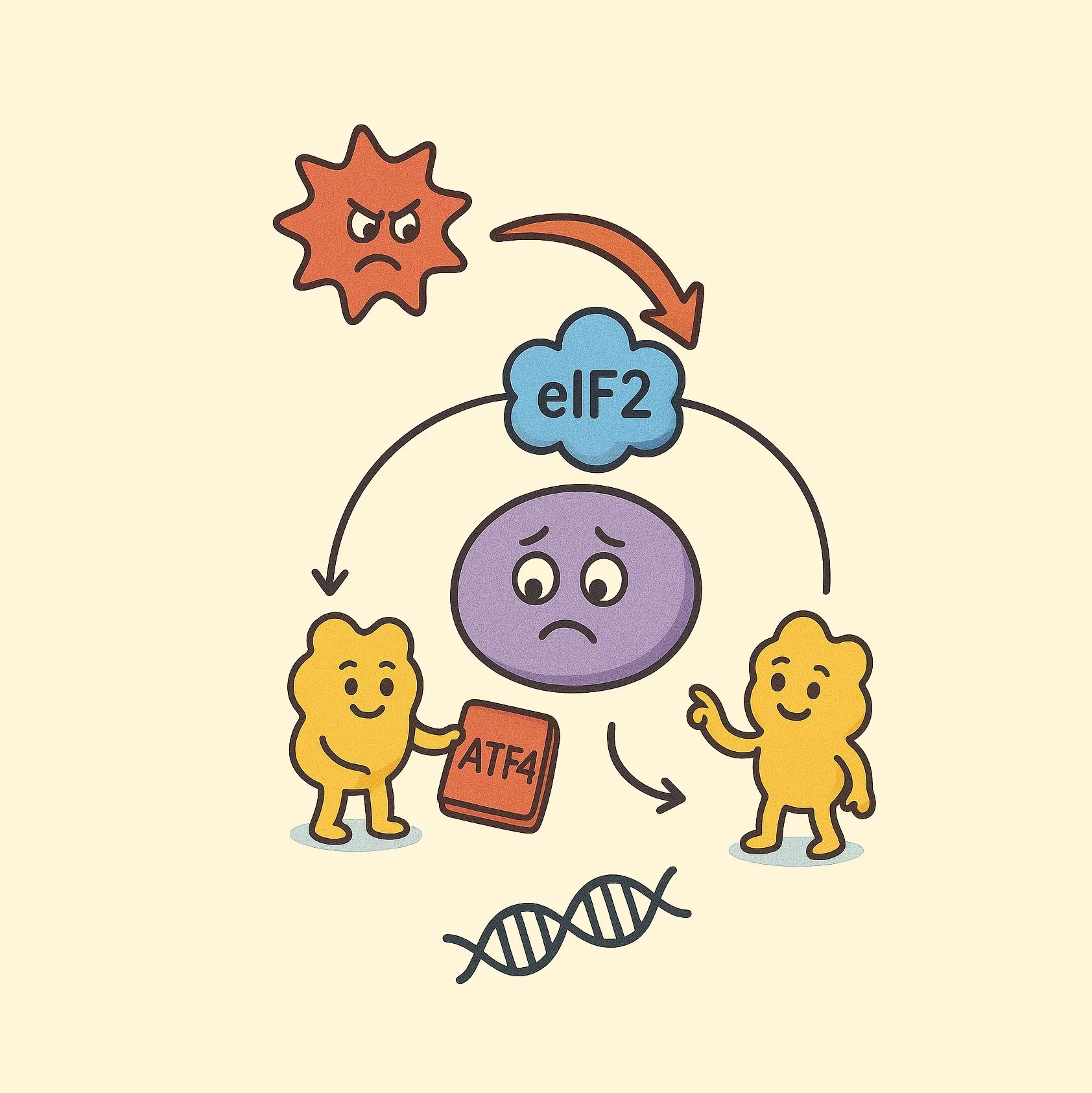
Proposed model for GCN2 activation and regulation of translational repression in response to collisions
Cancer cells sustain hyperproliferation through heightened protein synthesis and nutrient scavenging, depleting resources from the surrounding tumor microenvironment. This elevated translational demand creates unique dependencies on the protein synthesis machinery, presenting exploitable therapeutic vulnerabilities. We are focused on understanding how cancer cells mitigate the stress caused by their high protein production, with the goal of disrupting these survival mechanisms for therapeutic benefit. Specifically, we are examining: (1) how Integrated Stress Response (ISR)-mediated translational repression contributes to resistance in aggressive cancers subtypes, and (2) translational quality control failures generate immunogenic protein products that could be exploited for novel immunotherapy approaches These studies aim to define critical nodes where proteostatic regulation intersects with immune recognition in malignant cells.
Mechanisms that enable cells restore proteostasis following translational stress
We are investigating the cellular signaling networks and proteostasis surveillance mechanisms activated in response to proteotoxic stress. Our research focuses on elucidating how these various pathways coordinate protein degradation and adaptive gene expression to maintain proteome integrity. By delineating how these pathways regulate cell fate responses, we aim to uncover molecular determinants of cellular resilience. This knowledge has direct translational implications for therapeutic intervention in neurodegenerative and age-associated disorders rooted in protein misfolding and proteostasis imbalance